Adhesion / Extracellular Matrix
The Inner Life of the Cell | Harvard University, XVIVO
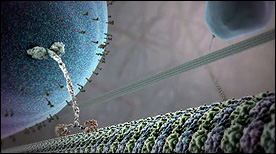
This Siggraph award-winning animation depicts the molecular players and signaling processes underlying leukocyte migration, adhesion and extravasation. Structural components of the cytoskeleton and the extracellular matrix, in particular, are highlighted.
» View the animation at Harvard BioVisionsCell Invasions | Charles Lumsden, Donald Ly, Jason Sharpe

This Maya animation provides a visual simulation of fibroblasts moving through extracellular matrix – the 3D matrix and behavior of the cell population through the matrix are based on mathematical models implemented in MEL.
» View the animationBack to top
Angiogenesis / Metastasis
Metastatic Bone Pain | Geoffrey Cheung
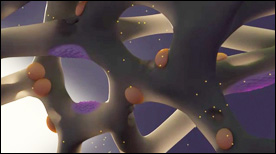
This animation shows the underlying peripheral pain mechanisms resulting from metastatic bone cancer.
» View the animationAngiogenesis | Drew Berry
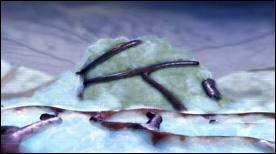
This animation shows the process by which tumors recruit new blood vessels thereby facilitating the metastatic behavior of stray cells that enter the circulation.
» View the animationBack to top
Apoptosis
Apoptosis | Drew Berry
This stunning Maya animation covers the death receptor signaling pathway that originates with binding of the Fas/TNF family of ligands, triggering of the caspase cascade, cytochrome C release from the mitochondria, apoptosome activation, and ensuing signal amplification.
» View the animationBack to top
Cell Division / Cell Cycle
Protein Expressions – Study N3 | Monica Zoppe, Scientific Visualization Unit
“The video PROTEIN EXPRESSIONS – Study N. 3D is the third (and last) re-elaboration of the movie on which we have been developing our studies in the last two years.”
» View the animationBack to top
Chemistry / Organic Synthesis
Si(111) Surface 7×7 Reconstruction | Yan Liang
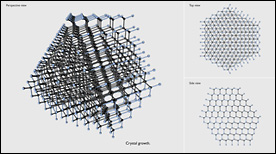
The Si(111) 7×7 reconstruction was one of the most intriguing problems in surface science. It took surface scientists over 25 years to determine its structure. This 4-minute long animation tries to help the viewers understand and enjoy the beauty of this complicated surface structure.
» View the animationDiversity Oriented Synthesis | Eric Keller
A step-by-step depiction of a diversity-oriented organic synthesis reaction on beads (created for Professor Stuart Schreiber at Harvard/Broad). At the same time as the camera follows the reaction in 3D showing bond rearrangements, the viewer can simultaneously follow the reaction in standard stick notation at the bottom of the screen.
» View the animation at National Institute of General Medical Sciences - NIGMSBack to top
Cytoskeleton / Molecular Motors
Fantastic Vesicle Traffic | Daniel von Wangenheim

How do plant cells deliver new material to their growing root hair tip? A combination of time-lapse photography, fluorescence live cell imaging and animation shows the molecular mechanisms in the context of the entire organism.
» View the animationMyosin Mechanism | Graham Johnson
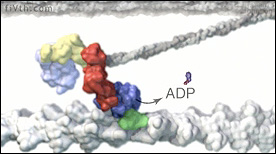
This animation describes the translation of chemical bond energy of ATP into the sliding motion of thick/thin filaments in our muscle fibers.
» View the animationMicrotubules: Structure, Function & Dynamics | Geordie Martinez, Steve Davy, Stylus Visuals
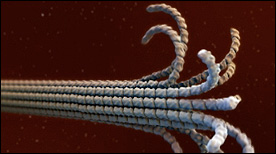
This Maya animation depicts the dynamic self-assembly and dissassembly processes of microtubules. The animation incoporates atomic resolution structural information for tubulin (as it undergoes a GTP vs GDP-induced conformational change), as well as cryoEM data for ‘protofilament peels’ and ‘helical ribbons’ from the Nogales lab.
» View the animationKinesin Mechanism | Graham Johnson
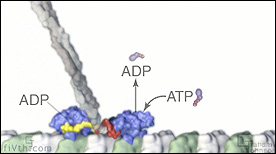
Kinesin walking along a microtubule protofilament demonstrating how energy exchanges combine with binding events to create forward motion.
» View the animationCell Quakes – Actin & Actinin | Anthony Zielinksi, Charles Lumsden

This movie presents a simulation of the behavior of selected cytoskeletal components as external forces are applied to the model (representing the forces of cell migration).
» View the animationBack to top
Developmental Processes
Embryonic Germ Layers | Biointeractive.org, Blake Porch, HHMI
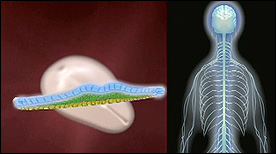
This animation briefly summarizes the early stages of development and highlights/maps the organ systems in the adult that result from the 3 embryonic germ layers.
» View the animation at HHMIEmbryonic Development | Biointeractive.org, Blake Porch, HHMI
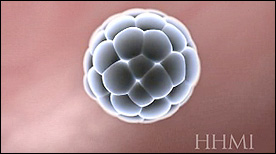
Covers the early stages of embyronic development (including fertilization, cleavage, blastocyst formation, implantation, cell migration in the inner cell mass and formation of the embryo’s germ layers and neural tube formation).
» View the animation at HHMIChick Embryo Development | AXS studio
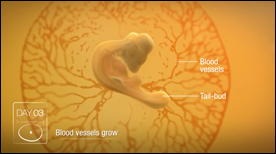
Animation of the 21 day development of a chicken embryo in the egg.
» View the animationBack to top
Disease / Immune System
Clostridium tetani and Tetanus | Maja Divjak

This animation describes the process of infection with Clostridium tetani and how the toxin it releases interrupts nervous control of our muscles, leading to tetanus.
» View the animationBordetella pertussis & Whooping Cough | Maja Divjak
This animation opens with normal respiratory function, then demonstrates how a bacterium, Bordetella pertussis, causes whooping cough by damaging ciliated airway cells.
» View the animationDiabetes (type I) | Etsuko Uno
This animation explores insulin production both in the normal case as well as in the development of Type 1 Diabetes, when insulin production is disrupted.
» View the animationMultiple Sclerosis | Gardenia Gonzalez Gil, Living Pixels
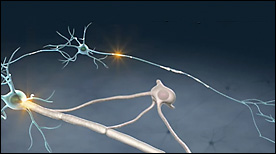
This Maya animation describes some immunological and brain barrier defects found in patients with Multiple Sclerosis. It illustrates how these defects progressively deteriorate neuronal signal transmission.
» View the animationCrohn’s Disease | Gardenia Gonzalez Gil, Living Pixels

The first two parts of this animation illustrate features of innate and adaptive immunity relevant to Crohn’s disease. The third part describes the mechanism of action of lipoxin resolving infection and inflammation, leading to restoration of healthy gastrointestinal function.
» View the animation at Living PixelsClonal Selection Theory | Etsuko Uno
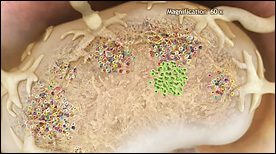
‘Fighting Infection by Clonal Selection’ was created to commemorate the 50th anniversary of a revolutionary theory called ‘Clonal Selection’ by Nobel Laureate, Sir Frank Macfarlane Burnet. The animation shows how clonal selection works during a bacterial infection of the throat.
» View the animationCancer is Not One Disease | Kate Patterson
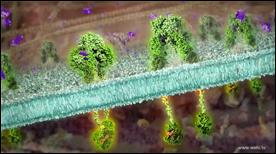
This animation draws on the experience of a patient living with pancreatic cancer to show some of the complexities of cancer. Over a long period of time, thousands of molecular ‘mistakes’ can accumulate in a cell and increase the risk of cancer. Now, these specific mistakes can be detected in individual cancer patients, thereby offering new hope for the future of personalised treatments.
» View the animationInsulin Receptor & Type II Diabetes | Maja Divjak
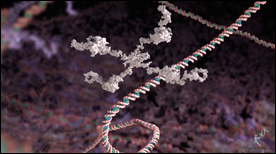
This animation describes the role of the insulin receptor in type 2 diabetes. It focuses on the very recent discovery of how the hormone insulin actually binds to the receptor on the surface of cells, as determined by Professor Mike Lawrence’s laboratory at the Walter and Eliza Hall Institute.
» View the animationBack to top
DNA / Chromatin
The Structure of DNA | Betsy Skrip, Sera Thornton
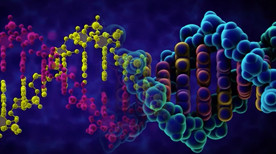
An exploration of the structure of deoxyribonucleic acid, or DNA.
This video was created for MITx 7.28.1x Molecular Biology: DNA Replication & Repair, offered on edX.
» View the animation at MITx BIOIntroduction to Epigenetics (for the Boston Museum of Science) | Digizyme
This animation explains how environmental conditions can influence the expression genes. We begin with a simplified view of gene transcription, mRNA synthesis and translation of the corresponding protein. We also observe how epigenetic ‘tags’ (chemical modifications to the DNA) can alter whether or not a gene is expressed.
» View the animation at DigizymeRestriction Endonuclease Digestion & Ligation | Drew Berry
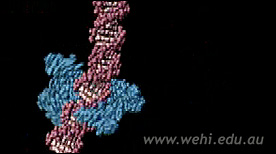
This animation depicts the proces of DNA recombination. The DNA plasmid is first digested with the restriction endonuclease enzyme ecoRI. Then, a piece of DNA encoding a gene is inserted into the plasmid by DNA ligase.
» View the animation at WEHIDNA structure | Drew Berry

A series of short animations highlighting the structureand flexibility of the DNA double-helix.
» View the animation at WEHIChromatin | Drew Berry
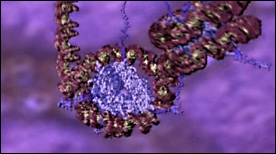
This animation shows the different levels of chromatin packing – starting with wrapping of DNA around histone octamers and nucleosome assembly, all the way to chromosome condensation during mitosis.
» View the animation at WEHIBack to top
Drug / Mechanism of Action
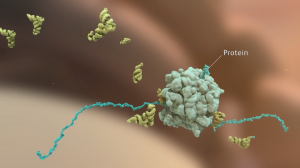
Penicillin and Antibiotic Resistance | Maria Voigt
This animation provides a detailed explanation for how penicillin and penicillin-related antibiotics disrupt the bacterial life cycle, as well as the underlying molecular mechanisms by which bacteria develop resistance.
» View the animation at RCSB PDBRapamycin, FKBP12 & FRAP | Biointeractive.org, Eric Keller
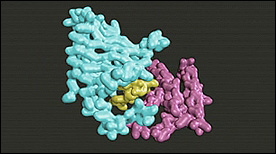
Dimerization of FKBP12 & FRAP is shown upon binding of the small molecule rapamycin.
» View the animationDNA Nanorobot (Wyss) | Digizyme
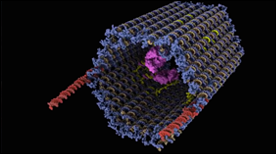
This animation depicts the structure and function of a drug delivery device created entirely of DNA. The therapeutic payload (in this case antibodies) is protected from the environment until a target protein (in this case PDGF, in green) competes for binding of the aptamer latches and triggers opening of the clamshell-like device.
» View the animationBack to top
Evolution / Origins of Life
Vesicle Entry of Adenosine Mono-Phosphate | Janet Iwasa
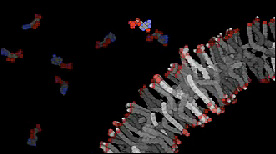
Polar molecules such as AMP may enter fatty acid vesicles through interactions between the fatty acid head groups and the small molecule.
» View the animationFatty Acid Vesicle Dynamics | Janet Iwasa
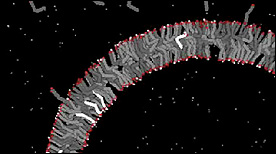
Although the vesicle structure itself as a whole is extremely stable, individual fatty acids within vesicles are extremely dynamic and are constantly joining and leaving the vesicle membrane. Protonated fatty acids (shown by the glowing hydrogen in the head group and the lighter colored tail) readily flip between the inner and outer leaflets of the membrane.
» View the animationFatty Acid Vesicle Formation | Janet Iwasa
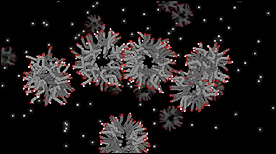
De novo vesicle formation from fatty acid micelles – Protons are represented by the small glowing spheres. Upon protonation, the micelle structure becomes more fluid and may allow for larger numbers of micelles to join together. Vesicle formation occurs by chance after the fatty acid sheet has reached a threshold surface area.
» View the animationFatty Acid Formation in a Geyser | Janet Iwasa

This animation illustrates a theoretical means by which fatty acids may have been synthesized along the sides of mineral walls of hydrothermal vents or (in this case) a geyser.
» View the animationBack to top
Metabolic / Respiration
How Enzymes Work | Maria Voigt
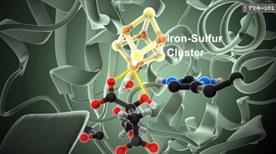
This animation describes how enzymes maintain the rate of thousands chemical reactions within a cell necessary for its survival, such as metabolism, protein synthesis, cell renewal, and growth. In particular, we see how aconitase is an enzyme of the citric acid cycle that uses its amino acid residues to catalyze a reaction.
» View the animation at RCSB PDBVillus Capillary – Hemoglobin | Gaël McGill
An animation that takes the viewer from the tissue level (i.e. a capillary inside a gut villus) all the way to the molecular level (by taking a look at the conformational changes that occur as a result of oxygen release by hemoglobin).
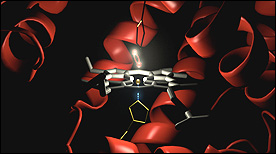
» View the animationSickle Cell Hemoglobin | Drew Berry
This animation depicts hemoglobin molecules binding to oxygen. The mutant form of hemoglobin is also shown and results in the assembly of the long stiff protein fibers characteristic of the disease sickle cell anemia.
» View the animation at WEHIHemoglobin | Janet Iwasa
A series of short movies decribing the structure of hemoglobin and the conformational changes that accompany binding of oxygen. Page also includes other useful resources.
» View the animationATP Synthase – Part I | Said Sannuga
The rotary mechanism of mitochondrial ATP synthase.
» View the animation at MRCATP Synthase – Part II | Said Sannuga
View from above and then below the F1 domain along the rotating γ-subunit.
» View the animation at MRCATP Synthase – Part III | Said Sannuga
How the rotating γ-subunit imposes the conformational states on a β-subunit required for substrate binding, ATP formation and ATP release.
» View the animation at MRCATP Synthase – Part IV | Said Sannuga
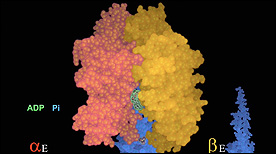
Three conformations of a catalytic β-subunit produced by 120º rotations of the central γ-subunit.
» View the animation at MRCATP Synthase – Part V | Said Sannuga

Changes in the positions of sidechains in the catalytic site of F1-ATPase bringing about binding and subsequent hydrolysis of ATP.
» View the animation at MRCATP Synthase | Graham Johnson

This animation describes the transfer of chemiosmotic energy into rotational energy, and ultimately into the chemical bond energy of ATP.
» View the animationBack to top
Neuronal Signaling
An Atomic View of Brain Activity | Burkhard Rammner

A 3D molecular-scale computational model of a synapse based on a combination of electron microscopy, super-resolution microscopy and biochemical data.
» View the animation at ScienceThe Whole Brain Catalog | Drew Berry
A visualization of the possibilities of the Whole Brain Catalog (http://wholebraincatalog.org), an open source, multi-scale virtual catalog of the mouse brain.
» View the animationNeural Long Term Potentiation (LTP) | Jason Raine
A 3D animation depicting the early molecular events underlying long term potentiation in the spinal cord of pain pathways. (Click on the icon in the “Master’s Research Project Examples 2002-2005 area of the page).
» View the animation at U. TorontoAlzheimer’s Enigma | Christopher Hammang
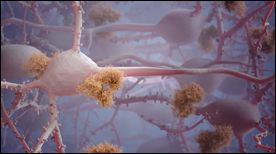
A beautiful 3D animation that takes us deep into the brain to understand how plaque build-up on brain cells occurs during Alzheimer’s disease.
» View the animationBack to top
Other
Entomology Animated Episode 1: RIFA Madness | Eric Keller

A stunning and humorous animation that explains the venom of the infamous fire ant species Solenopsis invicta.
» View the animationEntomology Animated Episode 2: “Chemistry Must Be Respected!” | Eric Keller

An entertaining animation demonstrating the chemistry of the amazing defense mechanism of the Bombardier Beetle.
» View the animationBack to top
Physiology
Heart simulation | Supercomputational Life Science & Sciement
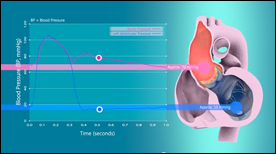
A highly realistic heart simulation that models heart function, contraction and relaxation of muscle based on the statistical behavior of molecules, which in turn are strongly coupled with the computation of blood flow and pressure.
» View the animationBack to top
Prokaryotes
E. coli Cytoplasm Brownian Dynamics Simulation | Adrian Elcock, Sean McGuffee
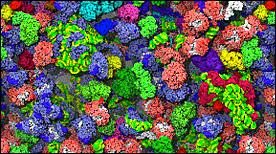
The simulation shows 1000 individual macromolecules diffusing, colliding and transiently associating with each other over the course of 10 microseconds of simulation; the translational diffusion coefficient of the GFP in this model is in agreement with experimental measurements.
» View the animationBacterial Flagellum | ERATO
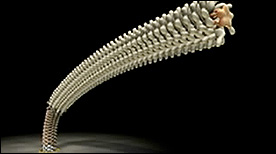
This series of animations depicts the processes of flagellar motion and assembly in molecular detail (also called the “Protonic Nanomachine Project”).
» View the animationBack to top
Protein Folding & Stability
What is a Protein? | Maria Voigt
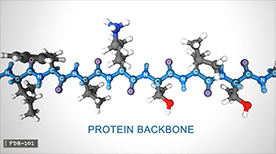
This animation from the Protein Data Bank guides us through the world of protein, which play countless roles throughout the biological world… from catalyzing chemical reactions to building the structures of all living things. Despite this wide range of functions all proteins are made out of the same twenty amino acids, but combined in different ways. The way these twenty amino acids are arranged dictates the folding of the protein into its primary, secondary, tertiary, and quaternary structure. Since protein function is based on the ability to recognize and bind to specific molecules, having the correct shape is critical for proteins to do their jobs correctly.
» View the animation at RCSB PDBSimulated ‘time-lapse’ of a nascent peptide | Barth van Rossum, Chris Spronk
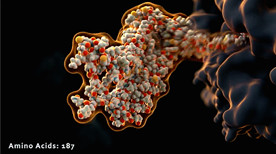
A beautiful animation showing a steered trajectory from a coarse-grained (no water) Molecular Dynamics/MD simulation of a peptide folding upon exit from the ribosome. The trajectory was calculated in Yasara, imported and morphed with Molecular Maya (mMaya) and rendered in Maya.
» View the animation at Spronk StudioProteasome | Janet Iwasa
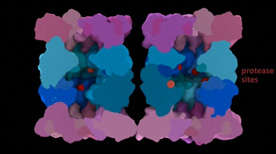
This Maya animation provides an introduction to proteasome structure as well as an explanation for proteasome-mediated degradation of a target protein (including potential “wobble” of the regulatory particle as it interacts with the core particle).
» View the animationProteasome & Ataxin | Biointeractive.org, Eric Keller, HHMI
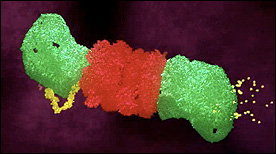
This Maya animation depicts the process of ubiquitin-dependent degradation in the proteasome. The effect of mutant ataxin no this process is also shown.
» View the animationFrom Milk to Yogurt | Yan Liang
A beautiful animation depicting how changing acidity conditions due to the presence of yogurt bacteria can affect the protein and fatty components of milk and induce formation of yogurt.
» View the animationBack to top
Replication
Tri Nucleotide Repeat | Biointeractive.org, Drew Berry, HHMI
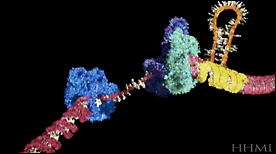
This animation shows how a tri-nucletoide repeat can cause the DNA polymerase to ‘slip’ and incorporate additional nucleotides during the replication process.
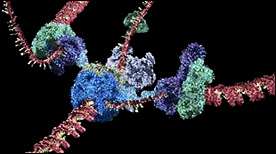
» View the animation at HHMIT7 Primase/Helicase | David Gohara, SciAna FilmWorks
This animation shows the dancing heptameric complex responsible for unwinding the DNA double helix in bacteriophage and how it is subsequently used as a site for primer synthesis.
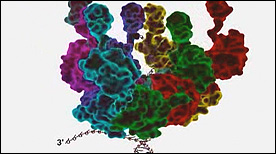
» View the animation at SciAnaFilmsReplication | Drew Berry
Still one of the more complex and beautiful molecular animations ever made, this movie shows the components and dynamic processes involved in the replication of both the leading and lagging strands of DNA.
» View the animation at WEHIBack to top
RNA Stability & RNAi
RNA Folding | Biointeractive.org, HHMI
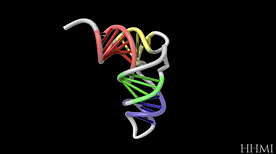
A short animated sequence showing how RNA can fold back onto itself (through the formation of intramolecular base-pairing).
» View the animationDicer | Geordie Martinez, Steve Davy, Stylus Visuals
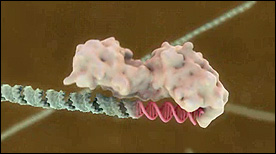
This Maya animation shows cleavage of double-stranded RNA into short RNA fragements by the Dicer ribonuclease.
» View the animationBack to top
RTKs & Signal Transduction
Clathrin Mediated Receptor Endocytosis | Janet Iwasa
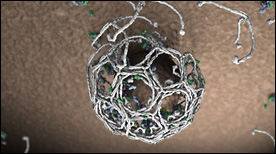
Dynamics of Lck in the T cell synapse – Upon T cell activation, clusters of signaling proteins form microdomains in the cell membrane. Some proteins, like the tyrosine kinase Lck (white) can freely diffuse between these clusters. Interactions between Lck and proteins in the signalling cluster can cause Lck to become immobilized.
» View the animationYeast Mating Pathway | Janet Iwasa
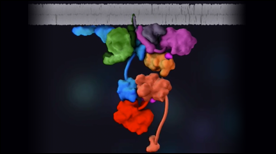
A 3D nimation depicting the step by step signaling transduction process of the mating pathway in yeast.
» View the animationBack to top
Stem Cells
The Origin of Breast Cancer | Drew Berry, Etsuko Uno
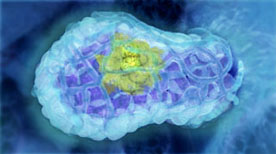
This animation visualises research published in Nature Medicine (Vol 15, Issue 8, 2009) by the laboratory of Jane Visvader and Geoffrey Lindeman. The mammary gland is comprised of three main cell types; alveolar, ductal and myoepithelial cells. Breast stem cells can develop into any of the three cell types through a series of intermediate cell stages. One intermediate is the luminal progenitor cell, which develops into either alveolar or ductal cells. The paper describes how an aberrant form of a luminal progenitor cell is involved in the development of some forms of breast cancer.
» View the animationThe Control of Breast Stem Cells | Drew Berry, Etsuko Uno
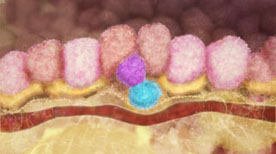
This animation illustrates how breast stem cells respond to steroid hormone despite the cells not having any steroid receptors. The animation illustrates the research published in Nature (Vol 465, Issue 7299, 2010) by the laboratory of Jane Visvader and Geoffrey Lindeman.
» View the animationBreast Stem Cells | Drew Berry, Etsuko Uno

An overview of the human mammary gland with a focus on the role of breast stem cells during pregnancy. The primary function of the mammary gland is to produce milk to nourish young offspring. The mammary gland is comprised of three main cell types; alveolar, ductal and myoepithelial cells. During pregnancy, the mammary gland increases in size due to the action of breast stem cells, which can mature into any of the three mammary gland cell types.
» View the animationStem Cell Introduction | Arkitek Studios
A series of animations with audio and text commentary that clearly explain the basics of stem cell biology (including their unique characteristics, pluripotency in the early embryo, presence in adult tissues and embryonic stem cells in culture).
» View the animationIntestinal Crypt Stem Cells – A Clonal Conveyor Belt | Digizyme, Eric Keller
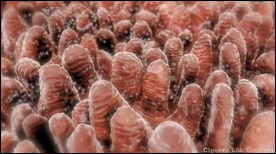
This animation, created for Hans Clevers’ lab, shows how the entire surface of the intestine is populated via a “clonal conveyor belt” mechanism. Daughter cells born from stem cells located at the base of the crypts travel up and differentiate, thereby pushing existing cells up towards the villus tip (the oldest cells are jetisoned via apoptosis at the villus tip). Adenoma formation is also shown.
» View the animationCSF Receptor | Drew Berry

A molecular view of the surface of a stem cell highlighting the binding of G-CSF by its receptor, dimerization, signal transduction and the resulting effect on cell division and growth.
» View the animation at WEHIBack to top
Transcription
PPAR Gamma transcription factor | Biointeractive.org, Eric Keller, HHMI
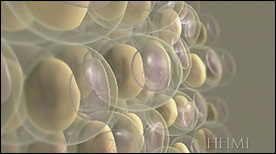
Shows fat cells in the adipose tissue adjacent to muscle – storage / breakdown of the cell’s fat droplet affects the balance of secreted adiponectin and resistin hormones. The effect of drugs against PPAR gamma is also shown to affect this balance and resulting insulin sensitivity.
» View the animation at HHMIPPAR Delta transcription factor | Biointeractive.org, Eric Keller, HHMI
Shows the effects of drug-binding to the PPAR-delta transcription factor receptor on DNA – a repressor is released thereby turning on the muscle delta network on genes. Oxidative metabolism is activated and leads to reduction of fat pads in adipose tissue.
» View the animation at HHMIp53 transcription factor | Biointeractive.org, Eric Keller, HHMI

This animation highlights the structure of p53 protein and its binding to a cognate promoter. Recruitment of RNA polymerase and transcription are also shown.
» View the animation at HHMIMECP2 transcription factor | Biointeractive.org, Eric Keller, HHMI
This animation show the effects of MECP2 DNA methylation (CpG islands) on recruitment of Sina3/HDAC, nuclesome modification and gene silencing.
» View the animation at HHMIDNA Central Dogma Part 1 – Transcription | Drew Berry
Transcription factors assemble at a DNA promoter region found at the start of a gene. Promoter regions are characterised by the DNA’s base sequence, which contains the repetition TATATA É and for this reason is known as the “TATA box”.
» View the animation at WEHIBack to top
Translation
tRNA-Ribosome Molecular Dynamics Simulation | K.Y. Sanbomatsu
One of the largest molecular dynamic simulations in biology – studies the interactions of tRNA as it enters the ribosome.
» View the animationTranslation | Drew Berry
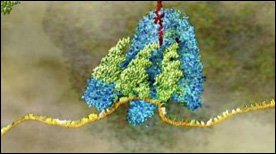
Part 2 in Drew Berry’s “Central Dogma” animations – the mRNA (yellow) is decoded inside the ribosome (purple and light blue) and translated into a chain of amino acids (red) as aminoacyl-tRNAs (green) deliver each amino-acid cargo (red/pink tip) to the ribosome.
» View the animation at WEHISignal Recognition Particle | Eric Keller, Steve Davy, Stylus Visuals
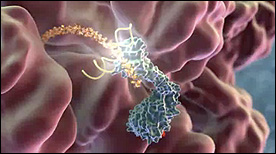
This Maya animation depicts the process by which the translating ribosome is halted by the signal recognition particle (SRP). The ribosome is subsequently brought to the membrane and docked with a channel to translocate the nascent polypeptide chain.
» View the animation at BloopatoneRibosome Function | Said Sannuga
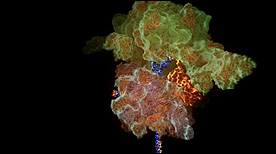
A detailed animation that covers all the central steps in prokaryotic translation (including initiation, elongation and termination steps with many of the individual protein factors involved in each).
» View the animationIRES | Stylus Visuals
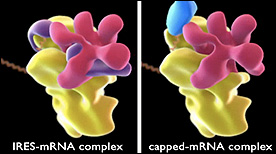
This animation compares the structure of ribosome complexes in either IRES-mRNA (Internal RIbosome Entry Sequence) or capped-mRNA conformations.
» View the animationGolgi / ER Visualization | Drew Berry
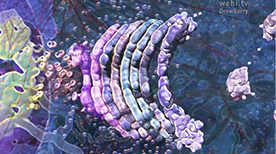
A visualization of a cell’s cytosplasm derived from electron tomography data from Brad Marsh’s laboratory. The different components – nucleus, microtubules, mitochondria, ribosomes, smooth ER, rough ER, Golgi – are highlighted in separate ‘passes’ and then overaid as one. A great reminder of how crowded cellular interiors are!
» View the animationElongation Factor Tu | Graham Johnson
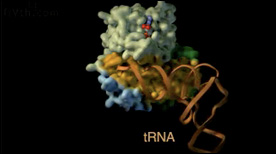
An animation highlighting the structural domains of elongation factor Tu and the surface involved in tRNA binding. The conformational change in the switch helix that occurs as a result of GTP hydrolysis results in the release of the tRNA.
» View the animationElongation Cycle of Protein Biosynthesis | A.H. Whiting, J. Frank, R. Agarwal
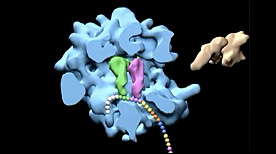
This visualization rotates the assembled ribosome and then shows (using a cut-away) the path of entry of the tRNA during the elongation cycle.
» View the animationBack to top
Viruses / Infectious Disease
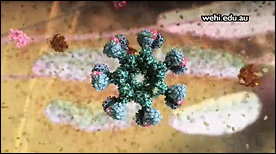
Model of Membrane Fusion by SARS CoV-2 Spike Protein | Gaël McGill, Jonathan Khao
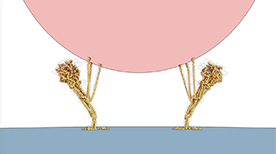
A dynamic simulation and visual model of how the SARS CoV-2 Spike protein drives fusion of viral and cell membranes. Binding of the S1 spike trimer ‘cap’ to an ACE2 receptor triggers release of S1 and unveils the S2 trimer below. Extension of S2 leads to embedding of hydrophobic fusion regions into the host cell membrane. Once tethered to both viral and cell membranes, refolding of S2 on itself drives membrane fusion and release of the viral genome into the host cell cytoplasm.
» View the animationHIV Life Cycle | Janet Iwasa
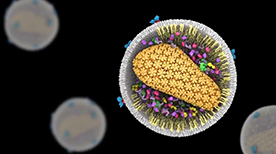
How does HIV infection occur? This molecular animation depicts the process of how HIV infects a T cell and transforms the cell into a viral factory.
» View the animation at Science of HIVA Molecular View of HIV Therapy | Maria Voigt
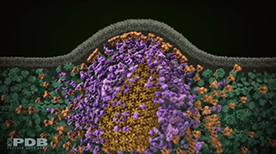
After HIV enters a T-cell, three enzymes play essential roles in the life cycle of the virus. Reverse transcriptase copies the viral RNA genome and makes a DNA copy. Integrase inserts this viral DNA into the cell’s DNA. In the last steps of the viral life cycle, HIV protease cuts HIV proteins into their functional parts. This animation from the Protein Data Bank describes the key events and molecular actors in HIV’s complex choreography.
» View the animation at RCSB PDBViral DNA Packaging – Part I | Eric Keller, Stylus Visuals
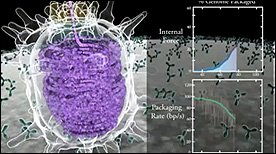
This 2-part Maya animation depicts the process of nucleic acid packing/assembly into the viral capsid. Part I shows the process simultaneous with the measured kinetics of packing and force (displayed on the right).
» View the animationViral DNA Packaging – Part II | Eric Keller, Stylus Visuals
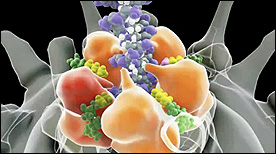
This 2-part Maya animation depicts the process of nucleic acid packing/assembly into the viral capsid. Part II focuses on the molecular machinery responsible for pulling the nucleic aacid strand inside the capsid.
» View the animationTomato Bushy Stunt Virus | Art Olson
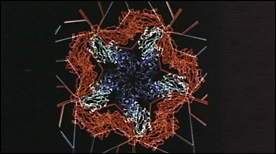
This 1981 landmark molecular animation was programmed in GRAMPS and captured from a computer screen with a Bolex 16 mm movie camera. Elegantly choreographed and paced, the movie presents the structure of the tomato bushy stunt virus (TBSV) – the first viral structure solved at atomic-resolution (2.9 angstroms) by Steve Harrison in 1978. Morphing animations of capsid proteins are also shown and explain the swelling of the viral particle observed at high pH.
» View the animationEarly Events in Reovirus Entry | Gaël McGill, Janet Iwasa
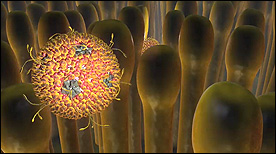
A more in-depth look at the early events of reovirus entry. This current version highlights each of the 8 proteins that make up the virus as well as its icosahedral symmetry. The virus is activated upon trypsin ‘attack’ and cleavage of the outer protein layer. The virus then binds to and enters the cell via the JAM-1 receptor and clathrin-mediated endocytosis.
» View the animationPseudomonas | Graham Johnson
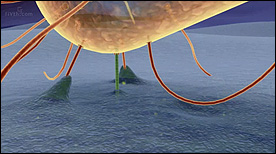
A sequence depicting Pseudomonas infection of lung epithelial cells.
» View the animationThe Lifecycle of Malaria (Part 1) | Drew Berry
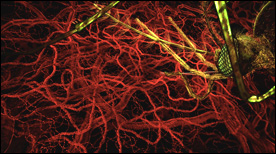
This animation represents part-1 of a 2-part series depicting the events of the malaria parasite lifecycle.
The parasite is shown entering the human host following a mosquito bite and we follow its progression initially to the liver and subsequently targeting erythrocytes on a large scale.
» View the animationThe Lifecycle of Malaria (Part 2) | Drew Berry
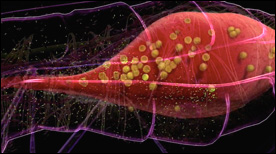
Part 2 depicts events in the mosquito host. The malaria parasite is shown reproducing in the mosquito’s stomach followed by the development of cysts and infection of the salivary glands.
» View the animationPoliovirus | Art Olson

This 1985 animation (programmed in GRAMPS) describes the structure of the poliovirus (seen here at near-atomic resolution). Icoshedral symmetry of the capsid, positioning and interaction of each of the V 1 – 4 proteins is described in detail.
» View the animationHIV Entry – gp41-mediated membrane fusion | Gaël McGill
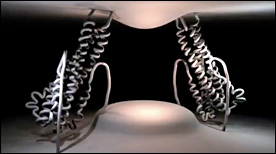
This Maya animation depicts the process by which HIV’s gp41 protein mediates the fusion of viral and cellular membranes during virus entry. In addition, strategies for inhibiting this process with peptide or small molecule inhibitors are shown.
» View the animationDengue Virus Entry | Digizyme, Gaël McGill, Janet Iwasa, Michael Astrachan, XVIVO
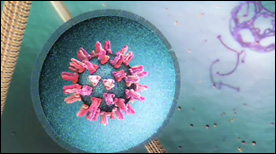
A narrated animation depicting the events that lead to Dengue virus entry into a host cell. In particular, rearrangements and conformational changes in the Dengue glycoprotein E are shown. These lead to membrane fusion and subsequent release of the viral payload into the host cell cytoplasm. Created for WGBH.
» View the animationCapsid Molecular Dynamics Simulation | Geordie Martinez, Stylus Visuals
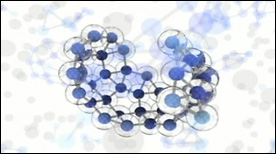
A Maya-rendered visualization of a VMD molecular dynamics simulation. Created for David Chandler’s lab at UC Berkeley, this movie depicts the physics of viral capsid formation while summarizing some of the technical steps involved in its creation.
» View the animationBacteriophage T4 | Seyet
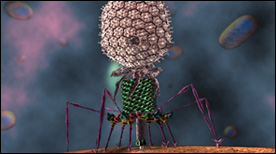
An accurate visualization of the Bacteriophage T4 based on Cryo-EM datasets of the virus. The scope of the animation is to show the infection process of T4 into an E. coli cell. All scientific data sets and motion based off of research from Michael Rossmann Laboratory (Purdue University).
» View the animation
Back to top




































































































